|
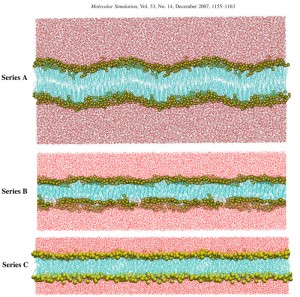
Surfactant bilayers: Surfactants, i.e. molecules consisting of a polar head and a long alkyl tail, generate quite different self-assembled structures, from micelles to lyotropic liquid crystalline ones. The later is composed of individual lamellae of fluid amphiphilic molecules separated by a solvent. Surfactant bilayers have been intensively studied for many years, and can be considered as a simple model for biological membranes. It should be noted that surfactants have been found already a wide range of applications in everyday life, in detergents, cosmetics, pharmaceuticals, food processing, agrochemicals, paints, paper coatings, etc. By using well-known ionic surfactant molecules sodium dodecyl sulfate (SDS) and sodium pentadecyl sulfonate (SPDS), we have investigated the lyotropic liquid crystalline phases of both molecules. A series of long 130ns molecular dynamics simulations of SDS/water system (512 SDS/15.000 water) were done using all atom and the united atom models. As software we use both NAMD and GROMACS codes with CHARMM27 and modified GROMOS87 force fields, respectively.
It has been stated that the CHARMM force field with all atom model gave the best results in comparison to the experimental findings, i.e. we found a strong agreement with the experimental values of area per molecule and hydrocarbon tail packing parameters. The above mentioned job was jointly done with our partners from the Institut für Chemie (Universität Potsdam) inPotsdam. The next step was the detailed study of long chain alkyl sulfonate/water system in lamellar phase, where as a surfactant SPDS molecule was used. In this regards, about 600ns parallel molecular dynamics simulation study was conducted for SPDS/water system. As the starting configuration was random, we examined the mechanism of self-assembly of SPDS molecules, i.e. we have explored in detail the dynamics of bilayer formation, which is indeed quite interesting as related to the dynamic self-assembly pathway leading to lamellar phase. Another interesting phenomenon was archived using the simulated annealing treatment. The temperature changing leads to the formation from the gel to fluid like phase with fully disordered hydrocarbon chains, i.e. the system undergoes gel-to-fluid phase transition.
|
|
|
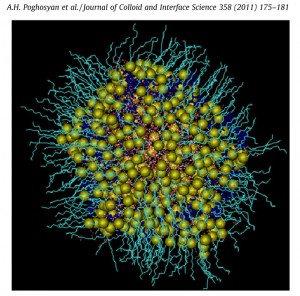
Inverse micelle/polyelectrolyte complexes: In collaboration with the Institut für Chemie (Universität Potsdam) in Potsdam, we have performed MD experiments of inverse sodium dodecyl sulfate (SDS) micelles in a mixed toluene/pentanol solvent in the absence and presence of a cationic polyelectrolyte, i.e. poly (diallyldimethylammonium chloride) (PDADMAC) aimed to estimate the influence of the PDADMAC on structural parameters of the microemulsion, such as the water droplet size, water properties or viscosity. The receiving MD data indicate a more rigid and ordered surfactant film due to the formation of a polyelectrolyte palisade layer in full agreement with the experimental findings, e.g. the viscosity increase and shift of the percolation boundary.
|
|
|
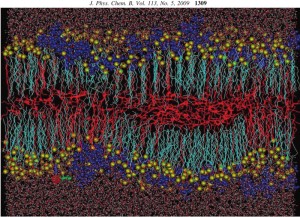
Surfactant bilayer/polyelectrolyte complexes: We have carried out a 50 ns of molecular dynamics study of PDADMAC/SDS/decanol/water systems, where the influence of the cationic polyelectrolyte on the anionic SDS-based lamellar liquid crystalline system was investigated. The polyelectrolyte-induced coexistence of two lamellar phases at a concentration of 2-3% of PDADMAC was observed, which is in agreement with experimental findings. The obtained MD results also provide us important information on the dynamical and structural features of the abovementioned complex system, as well as the conformational properties of the polyelectrolyte and the surfactant molecules become available. This work also was done in collaboration with the Institut für Chemie (Universität Potsdam) in Potsdam.
|
|
|
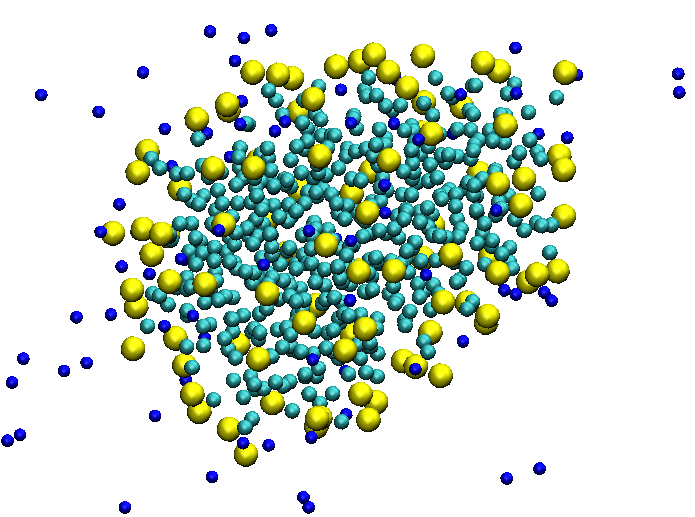
Coarse-grained simulations: The coarse-grained (CG) approach, which is originally developed by Marrink and coworkers, provides to simulate huge systems with long time periods. We tried to study the mechanism of the self-assembly of long alkyl tail ionic micelles using the so called “granular” approach. During last decade, a number of simulations were reported, where many authors claim that it is very difficult to reach an equilibrium using atomic scale MD, as it is known that the lifetime of micelle formation is an order of microseconds. In this regards, we have carried out CG simulations using GROMACS code with MARTINI force field. We have reached up to 80μs timescale and tracked final formation at 50μs timepoint. A key structural parameters, such as radius of gyration, characteristic relations of chain were estimated to understand the mechanism of self-organization process.
|
 |
|
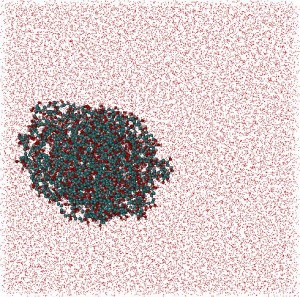 Polymers: In collaboration with Belarusian State Technological University, a MD and statistical-mechanics study of polyvinyl alcohol (PVA)/water and polyvinyl pyrrolidone (PVP)/water systems with weight ratio Cpolymer/Cwater = 1/4 was carried out. After 30ns of MD simulation for each system globular structures for both polymers were determined. Polymers: In collaboration with Belarusian State Technological University, a MD and statistical-mechanics study of polyvinyl alcohol (PVA)/water and polyvinyl pyrrolidone (PVP)/water systems with weight ratio Cpolymer/Cwater = 1/4 was carried out. After 30ns of MD simulation for each system globular structures for both polymers were determined.
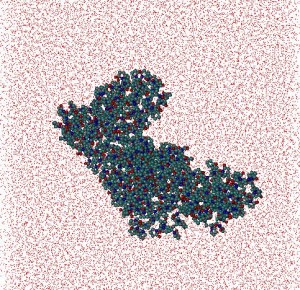 Some structural parameters such as polymer coil average radius, polymers radii of gyration, characteristic relations of polymer chains were estimated. Also the parameters describing the relative position of monomers were examined. Particularly, the static correlations between these parameters were investigated. It was obtained that the planes of separate monomers are chaotically distributed, while the angles between the nearest carbon bonds are approximately constant. Some structural parameters such as polymer coil average radius, polymers radii of gyration, characteristic relations of polymer chains were estimated. Also the parameters describing the relative position of monomers were examined. Particularly, the static correlations between these parameters were investigated. It was obtained that the planes of separate monomers are chaotically distributed, while the angles between the nearest carbon bonds are approximately constant.
Biological membranes
|
Pure phospholipid bilayers: The phospholipid bilayers have long been intensively investigated as the structural model for biological membranes and have attracted much attention. Dipalmitoyl phosphatidylcholine (DPPC) has been regarded as the benchmark lipid, since many real experiments (X-ray, NMR, etc) were done namely with this lipid, which have a dipolar hydrophilic head group soluble in water and two extended hydrocarbon tails. Our earlier jobs were concerned namely the detailed MD study of pure DPPC bilayers. We had performed an MD simulation of 128 DPPC/3655 water system in liquid crystalline state, thus constituting by weight (water molecules per lipid), which corresponds to a fully hydrated lipid bilayer. It was shown that at average the hydrogen atoms of water are oriented towards the phosphorus atoms and the negatively charged oxygen atoms – towards the nitrogen atoms caused by the large number of hydrogen-bonded groups, which may make important contribution to the intermolecular interaction. The properties of hydrocarbon chains and headgroup (order parameter, tilt angle, inclination of dipole fragment, peak-to-peak distance, etc) were determined and compared with literature data to evaluate its reliability and in overall the obtained MD results were in good agreement with existing experimental data.
|
|
|
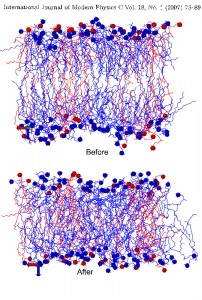
Mixed phospholipid complexes: Biological membranes are complicated multicomponent formations and their properties, such as ion transport, cell fusion, drug susceptibility and interaction with anesthetics depend on the physical properties and state of the membrane constitutes lipids and also the most important depend on the lipid composition of the local regions of the membrane. The lipid bilayer of biological membranes consists of the mixture of several lipids, i.e. a lipid component of any biological membranes are the mixtures of various type of lipids, having same common structure, but differing by the structure of polar part, and especially the hydrocarbon chains. In this regards, we have performed atomistic MD simulation of DMPC/DPPC mixed bilayers (same headgroup, differing only by hydrocarbon chain), consists of various fraction of lipids (25%, 50% and 75%). The aim of this work is to reveal the nature of mixing, e.g. in terms of chain orientational order and interdigitation of tail, taking into account that the interdigitation effect is important to explain in the ions and other compounds permeability through the membrane. The received MD data claims that the interdigitation degree of hydrocarbon chain is increased as the DPPC fraction is decreased. In case of racemic 50% mixture, the area per lipid value was in good agreement with experimental estimations. The DMPC/DPPC mixtures behave as almost ideally mixtures.
|
|
|
Erythrocyte model membranes: A MD simulation with duration of 180 ns of human red blood erythrocyte asymmetric model membrane has been successfully carried out. The construction of model membrane was implemented by using of MDesigner software by creating the molecules of POPE, SAPE, POPC, SOPC, SAPS, SDPS, LSM, HSM, and cholesterol (overall 256 phospholipids /8572 water molecules). The GpA protein transmembrane part were inserted into the system. The main structural parameters (area per phospholipid molecules on the membrane surface, membrane thickness, membrane surface roughness function, etc) have been calculated and compared with exiting experimental findings in order to verify the accuracy of MD simulations, paying particular attention to conformational dynamical features of embedded GpA protein by calculating the angle between GpA α and β helices, as well as exploring the immediate surroundings of the GpA in the membrane upper and lower parts.
|
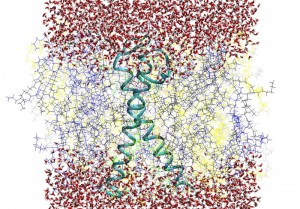
|
|
Membrane / Taurine interactions: The main goal is to create the model of human erythrocyte membrane as close to its natural biological state as possible. We have already created a bilayer consisting from these 5 different types of lipids. The steps would be to add to the membrane transmembrane protein and a taurine molecule on the surface of the bilayer in a water environment. The interactions between taurine molecule and the bilayer will be investigated. The project is going to be carried out by the method of computer experiment particularly by using molecular dynamics simulations. The GROMACS simulation package is being used by its default force filed. By getting this model we can measure various parameters of human erythrocyte membrane-taurine interaction and also make clear its many aspects even by visually showing them. After this measurements and analyses we can answer on many questions about taurine influence on the cell membrane which is still unclear and which can be a huge benefit for understating of some diseases’ mechanisms.
|
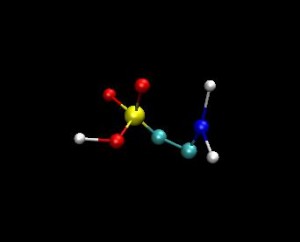
|
| |
|
|